Someone recently asked how to create a model where a molecular species received a regular dose at a fixed interval of 12 hours.
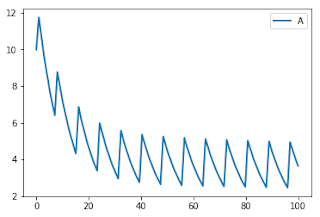
They wanted something where a species A is being degraded but at regular intevals is increased by a fixed amount. See the plot below.
Lucian Smith and I came up with two possible solutions which are shown below. The first solution from Lucian is to set up a variable called trigger that we define using the differential equation trigger’ = 1 (note the apostrophe). The ode ensures that trigger increases linearly during the simulation. We then set up an event that triggers when trigger is greater than 12 hours at which point trigger is set back to zero and we add the dose of value 3 to species A. trigger then starts to integrate again from zero and the cycle repeats.
import tellurium as te
r = te.loada("""
A ->; k1*A;
k1 = 0.1; A = 10
trigger = 0
trigger' = 1
at (trigger>12): A = A+3, trigger=0
""")
m = r.simulate (0, 100, 100, ['time', 'A'])
r.plot()
The second solution is less elegant but increases the trigger value when an event occurs. In the code below when time exceeds the current trigger value we add the dose then increase trigger by 12 hours so that can it can be triggered again.
m = r.simulate (0, 100, 100, ['time', 'A'])
r.plot()
r = te.loada("""
A ->; k1*A;
k1 = 0.1; A = 10
trigger = 12
at (time > trigger): A = A+3, trigger = trigger + 12
""")
m = r.simulate (0, 100, 100, ['time', 'A'])
r.plot()
No comments:
Post a Comment