October 20, 2017 10:31 am
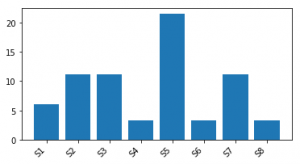
I had a model with 27 floating species and I wanted to plot the steady-state concentrations on a histogram where the labels were the names of the different species. Here is a general-purpose script that will do that:
def plotFloatingSpecies (r, width=12, height=6):
import matplotlib.pyplot as plt
xlabels = r.getFloatingSpeciesIds()
concs = r.getFloatingSpeciesConcentrations()
plt.figure(figsize=(width,height))
plt.bar(xlabels, concs, label=xlabels)
plt.xticks(range (len (xlabels)), xlabels, ha='right', rotation=45)
The function has a default width and height for the resulting plot. The default widens the usual size so that the labels don't collide. The first argument is the libroadrunner instance. To give a contrived example, the following is a model of 8 floating species that form a linear chain governed by simple irreversible mass-action kinetics.
import tellurium as te
import roadrunner
def plotFloatingSpecies (r, width=12, height=6):
import matplotlib.pyplot as plt
xlabels = r.getFloatingSpeciesIds()
concs = r.getFloatingSpeciesConcentrations()
plt.figure(figsize=(width,height))
plt.bar(xlabels, concs, label=xlabels)
plt.xticks(range (len (xlabels)), xlabels, ha='right', rotation=45)
r = te.loada("""
$Xo -> S1; k1*Xo;
S1 -> S2; k2*S1;
S2 -> S3; k3*S2;
S3 -> S4; k3*S3;
S4 -> S5; k4*S4;
S5 -> S6; k5*S5;
S6 -> S7; k4*S6;
S7 -> S8; k3*S7;
S8 -> ; k4*S8;
k1 = 0.3; k2 = 0.5; k3 = 0.27; k4 = 0.9; k5 = 0.14
Xo = 10;
""")
r.steadyState()
plotFloatingSpecies (r, width=6, height=3)
The resulting plot is shown below:
No comments:
Post a Comment